NATIONAL MUSEUM OF NATURAL HISTORY
Scientists to Read DNA of All Eukaryotes in 10 Years
Researchers at the Smithsonian and around the world are working to sequence the genomes of every eukaryotic species on Earth in the next 10 years through the Earth BioGenome Project.
/https://tf-cmsv2-smithsonianmag-media.s3.amazonaws.com/blogging/featured/A_nearly_complete_puzzle_of_a_blue_DNA_strand_on_black_background..jpg)
When Rosalind Franklin was born 100 years ago, scientists already knew that physical traits are passed from one generation to the next through chromosomes. They just didn’t know how.
When early geneticists picked apart chromosomes, they found mostly protein molecules and deoxyribonucleic acid, or DNA. They thought that the larger, more complex protein molecules were what stored the code for genes and life. It wasn’t until the 1940s and 50s that they began to consider DNA as the source of the genetic code.
By that time, Franklin — a chemist and expert in using X-rays to look at molecular structures at King’s College London — captured X-ray images of DNA. These images played a crucial role in discovering the structure of DNA.
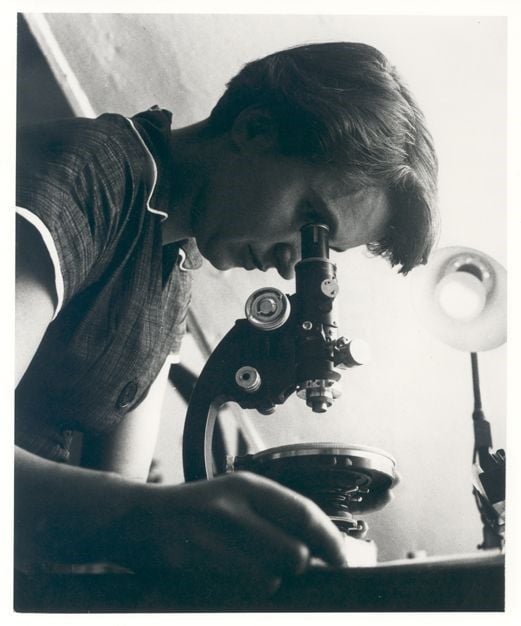
Since then, scientists have moved beyond piecing together what DNA molecules look like to reading the entire genetic code that it stores, called genomes, in thousands of organisms. But that number will quickly skyrocket into millions as researchers at the Smithsonian and around the world work to sequence the genomes of every eukaryotic species on Earth — that is, everything besides bacteria and archaea — in the next 10 years through the Earth BioGenome Project.
A powerful tool for problem-solving
Identifying and comparing the genes that give organisms their traits gives scientists a powerful tool for solving long-term problems. It helps researchers engineer better crops, find cures for genetic diseases, track how pathogens spread, manage pests, preserve endangered species and learn more about how life evolved.
“Every time we sequence something, whether pathogens, their hosts, or endangered species, we answer questions we didn’t even know we should be asking,” said Warren Johnson, a National Research Council research associate at the Smithsonian Walter Reed Biosystematics Unit.
Scientists expect the global sequencing effort to reveal previously unknown organisms. In addition to the almost two million eukaryotic species already known to science, we expect to find millions more, according to John Kress, botanist emeritus at the Smithsonian’s National Museum of Natural History. He’s one of three co-chairs of the Earth BioGenome Project.
“One of the best things about the Earth BioGenome Project is scientists getting together around the world to do this,” said Kress. “And, in the process, generating a lot of genomic data that will help us understand how nature works and how we can work better with nature.”
Museums play a new role
One of the major challenges in large genomic projects is getting access to well-preserved tissues that researchers need for extracting and sequencing DNA. Finding and preserving tissues from every organism would be costly and time-consuming.
About eight years ago, the National Museum of Natural History built a warehouse of freezers and liquid nitrogen tanks, called a biorepository, for storing tissues and DNA samples. Scientists around the world use these samples for sequencing — saving time, resources and trips to remote field sites.

“This has not typically been one of the missions of museums around the world,” said Jonathan Coddington, senior research entomologist at the Smithsonian’s National Museum of Natural History. “But museums have always been in the business of supporting current research.” Moving forward, more of that research will involve working with DNA.
Coddington leads the Global Genome Initiative — which supports a network of over 95 organizations across 30 countries working to collect and preserve Earth’s biodiversity in these types of biorepositories within six years. The Initiative’s collections and data will help researchers make the Earth Biogenome Project’s colossal goal a reality.
Cracking the code of life
Genomics studies have already disrupted much of what we thought we knew about the natural world. Researchers traditionally defined and grouped species together based on their appearance and physical traits. But once scientists began sequencing genomes, they realized some species are much more closely related than previously thought, while others are barely related at all.
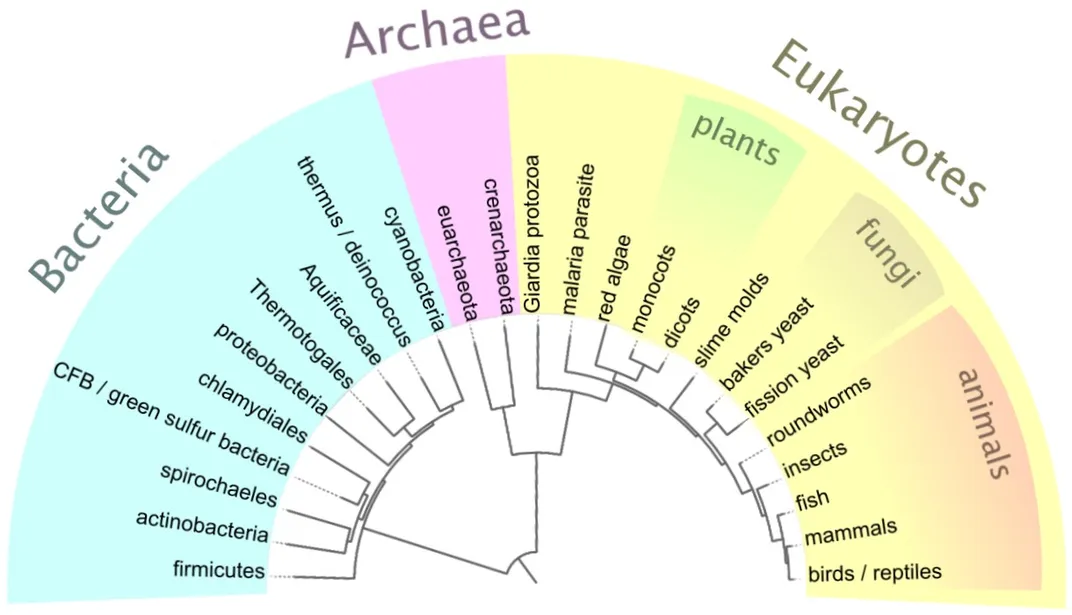
“It has completely reorganized what we understood about the evolutionary history of plants,” said Coddington. “The same thing happened in birds and in spiders. Group after group after group.”
Revealing how organisms evolved and relate to each other helps scientists understand how they adapt to changes. This knowledge can help humans adapt — through agricultural development, biomedical advances and even behavioral changes — to challenges like pandemics and global climate change.
“Genomics may well be the reason why we are able to make it through this coming period of global climate change in as healthy a way as possible,” said Johnson from the Smithsonian Walter Reed Biosystematics Unit. “It’s going to completely change the way that we see and interact with the world.”
Comparing genomes across species can also teach researchers about how organisms interact with each other. For example, scientists discovered that many parasites have lost some of their genes and instead rely on their hosts’ genomes. Some of them have extraordinarily short genomes, yet still manage to change and take advantage of their host’s behavior.
“I find it completely fascinating that something with a much simpler genome can manipulate an animal that is arguably much more complicated than itself,” said Katrina Lohan, a parasite ecologist at the Smithsonian Environmental Research Center. She added that efforts like the Earth BioGenome Project could help parasitologists identify unknown species, but additional research is required to understand their impacts on individuals and ecosystems. “We know so little about the diversity of parasites in general.”
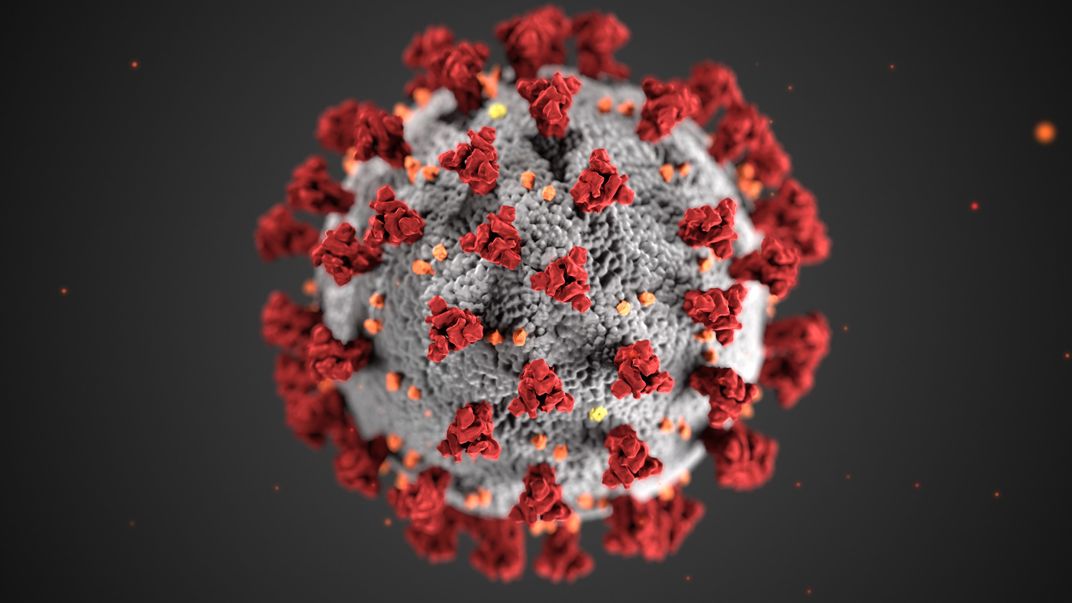
Researchers also study the spread of parasites and pathogens by examining the genomes of their hosts. SARS-CoV-2 — the virus responsible for the COVID-19 pandemic — enters human cells by tricking a particular protein, known as ACE2, that acts as a gatekeeper into letting it inside. A group of researchers at the Smithsonian and around the world recently compared the DNA and protein sequences of ACE2 of 410 animal species to find organisms with similar proteins that the virus might be able to trick. The scientists used available genomes to find the ACE2 sequences. They used this information to predict which animals besides humans might be susceptible to contracting and spreading COVID-19.
“I think it's a great example of the power of comparative genomics,” said Klaus-Peter Koepfli, a conservation biologist at the Smithsonian’s Conservation Biology Institute who worked on the project. He and other scientists who work with genomes expect our understanding of biology to expand in exciting ways as they unravel the secrets within DNA.
Related stories:
Viper’s DNA Reveals Ancient Map of South America
Safety Suit Up: New Clean Room Allows Scientists to Study Fragile Ancient DNA
Can Genetics Improve Fisheries Management?

