Google Releases A.I. That Can Predict How the Human Body’s Molecules Behave, Boosting Drug Discovery Research
Called AlphaFold 3, the latest update of the software models the interactions of proteins with DNA, RNA and other molecules for the first time
:focal(960x544:961x545)/https://tf-cmsv2-smithsonianmag-media.s3.amazonaws.com/filer_public/f1/c4/f1c4ab29-c6b6-4088-a04a-b804c783b0fb/deepmind.webp)
Last week, Google released a much-anticipated upgrade for its AlphaFold software, which harnesses artificial intelligence to predict the shape and structure of molecules within the human body.
Any given molecule’s shape is indicative of its function and behavior, so biologists have long researched how chains of amino acids, the building blocks of proteins, fold into various shapes. The A.I. tool can accelerate and streamline this process, opening new avenues for breakthroughs—notably in vaccine and drug development.
AlphaFold 3, the newest update from Google DeepMind described last week in the journal Nature, builds on its previous two iterations. The software’s initial tease in 2018 offered potential for accurately predicting the three-dimensional structure of proteins, while its 2020 update, AlphaFold 2, came with significant improvements. In 2021, Google released an open-source version of AlphaFold, along with the predicted 3D structures of nearly all known proteins in the human body. The next year, two million predicted protein structures were shared.
But despite these leaps forward—which helped researchers map the human heart and better understand extinct birds’ eggs—AlphaFold 2 was limited in scope to modeling proteins.
“The AlphaFold 2 system only knew about amino acids, so it was of very limited utility for biopharma,” Mohammed AlQuraishi, a systems biologist at Columbia University who is not affiliated with Google DeepMind, tells MIT Technology Review’s James O’Donnell.
/https://tf-cmsv2-smithsonianmag-media.s3.amazonaws.com/filer_public/8e/cf/8ecf8233-17b3-47fe-a71c-38637ee9d744/deepmind2.png)
The newest version of the software can predict not only the shape of proteins, but also the structures of DNA, RNA and other molecules, such as ligands. Crucially, this update will allow researchers to better predict and study how different molecules in the human body geometrically interact with each other—and anticipate where a drug might bind to a protein.
This ability can “save months of experimental work and enable research that was previously impossible,” Deniz Kavi, a co-founder and chief executive of Tamarind Bio, a drug discovery start-up, tells the New York Times’ Cade Metz. “This represents tremendous promise.”
Researchers could use the software update to probe some initial questions, including how proteins respond to DNA damage within the human body.
“It tells us a lot more about how the machines of the cell interact,” John Jumper, the director of Google DeepMind, tells the New York Times. “It tells us how this should work and what happens when we get sick.”
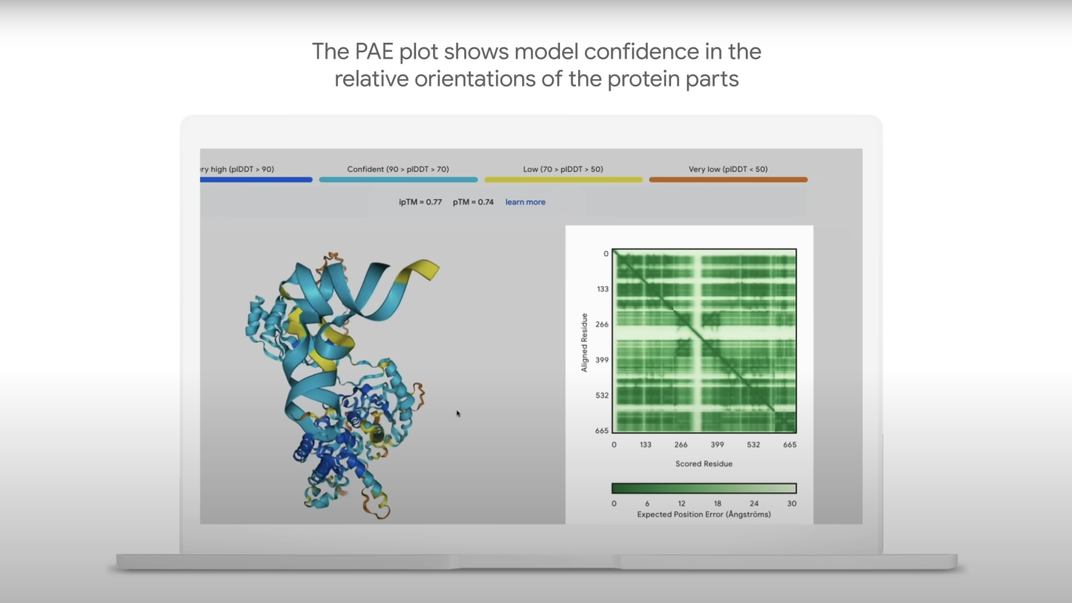
AlphaFold 3 offers researchers a level of confidence, often ranging from 40 percent to 80 percent, with each prediction it models, according to MIT Technology Review. Parts of a structure modeled with high confidence appear in blue, while the more uncertain regions appear in red. In some areas, its inaccuracy is a limitation—for modeling RNA-protein interactions, for example, the system isn’t yet highly exact.
Another potential drawback is the model’s ability to “hallucinate,” or produce false information. The team behind AlphaFold 3, in order to build its molecular library and function, borrowed methods from other A.I. models, such as OpenAI’s DALL-E 2 and Sora, that generate images and video. This improved AlphaFold 3’s capacity to produce large 3D images of molecular shapes, but leaves it liable to hallucinate. The team hopes to alleviate this issue with more training data, and in the paper, they note that hallucinated structures would typically be marked as low confidence.
Unlike its predecessor, the code for AlphaFold 3 will not be made open-source, and only a limited version, the AlphaFold Server, will be released publicly.
“This is a big advance for us,” Demis Hassabis, the CEO of Google DeepMind, tells WIRED’s Will Knight. “This is exactly what you need for drug discovery: You need to see how a small molecule is going to bind to a drug, how strongly and also what else it might bind to.”
/https://tf-cmsv2-smithsonianmag-media.s3.amazonaws.com/accounts/headshot/ChristianThorsberg_Headshot.png)

/https://tf-cmsv2-smithsonianmag-media.s3.amazonaws.com/accounts/headshot/ChristianThorsberg_Headshot.png)